
SMC proteins
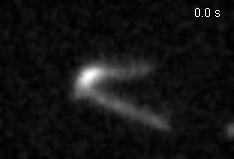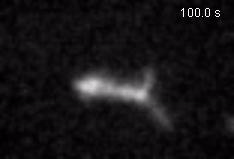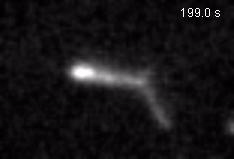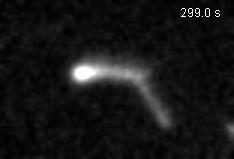
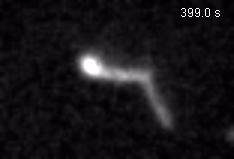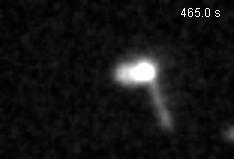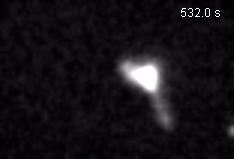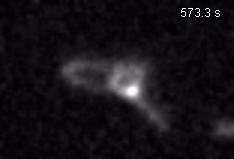
Chromatin loops are one of the basic structural units of three-dimensional chromosome organization. Loops were hypothesized to form by ‘loop extrusion,’ by which structural maintenance of chromosomes (SMC) complexes bind a chromatin fiber and reel it in, extrude it as a loop. Recently, we provided direct evidence of DNA loop extrusion activity of condensin SMC complexes (Fig.1) and their ability to bypass one another and generate higher-order loop structures (Fig.2). However, it still remains mysterious how these unique new classes of motor proteins carry out their loop extrusion activities at the molecular level. We hope to bridge this knowledge gap by obtaining hi-resolution structural information of the reaction intermediates via cryo-EM and cryo-electron tomography approaches.
Moving further, we are interested in investigating the roles of other SMCs and SMC-like (e.g. Rad50, RecN and Smc5/6) proteins in genome organization and maintenance.
References
[1] E. Kim, J. Kerssemakers, I. A. Shaltiel, C. H. Haering, C. Dekker, DNA-loop extruding condensin complexes can traverse one another. Nature 579, 438–442 (2020)
[2] M. Ganji, I.A. Shaltiel*, S. Bisht*, E. Kim , A. Kalichava, C.H. Haering, C. Dekker, Real-time imaging of DNA loop extrusion by condensin. Science 360, 102-105 (2018)

